Title: Reducting storage requirements for biological sequence comparision
Content
1.Motivation?
compute read overlap when do sequence comparision (including nucleic acid / protein / etc.), too much seeds need to be stored, but which cannot be stored in RAM.
the existing method’s catagories?
- “seed and extend” approaches, e.g. BLAST_1990
- “seeds” without an explicit extend step, e.g. SSAHA_2001
2.why minimizers?
Minimizers, only a small fraction fo all seeds.
- speed up string-matching comparision
- missing only a small fraction of the matches (using all seeds)
This is the fundamental reason why using minimizers (rather than all k-mers) as seeds reduces storage requirements.
3.how to select minimizers(a fewer representative kmers)?
Way 1.every k-th k-mer (each letter is covered exactly once) don’t work, because that. However, in that case two strings Ti and Tj with long identical subsequences that start at positions pi and pj need not have a stored k-mer in common unless pi − pj is a multiple of k. Thus, the database would not satisfy the collection criterion, in the sense that sorting it by k-mer would yield seeds for only a small fraction of matching pairs (Ti , Tj ).
Way 2.different strings Ti and Tj choose the same representative if they share a long enough subsequence. 选择原则是,对于不同的序列(Ti and Tj), 他们共有的长子串可以被同样的kmer所代表。这样被选出的kmer叫做minimizers。
4.minimizers’ properties?
- Property1. If two strings have as ignificant exact match,then at least one of the minimizers chosen from one will also be chosen from the other. (If two strings have a substring of length w+k−1 in common, then they have a (w, k)-minimizer in common.) 有相同子串,就一定会共享同一个minimizer
5.Flowchart of minimizer selection
Step1.select an ordering for the set of all k-mers
Step2.examine w consecutive k-mers and select the smallest as a minimizer. a set of w consecutive k-mers covers a string of exactly w + k − 1 letters, where ‘consecutive’ means that each k-mer is shifted by one letter from the previous one.
6.An example of minimizer
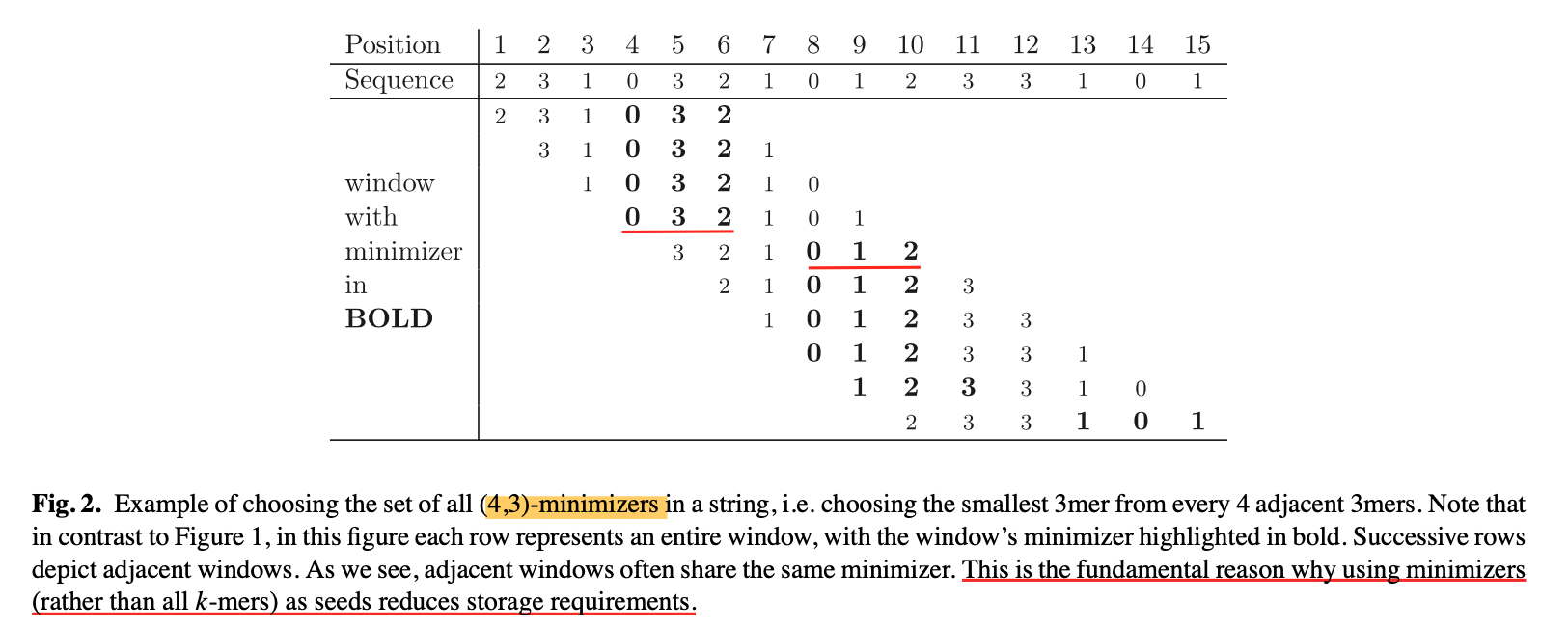
7.为啥要保证minimizer cover 所有bases 啊?
1). 保证minimizer cover 所有bases的策略:
- w ≤ k, every base in a string will be covered
- use (u, k)-end- minimizers, the ends of strings will be well covered (combine (w, k)-minimizers of a string with (u, k)-end- minimizers for u = 1,…,w − 1 at both ends of the string)
increasing the likelihood of finding low-fidelity matches on the ends of strings ? Properties 1′ and 2 imply that two strings with an exact overlap of at least 2k bases have a minimizer in common.
2). minimizer 没覆盖所有bases的情况(w ≫ k): ordering 顺序的选择更为重要,通过设置排序方式,尽量选到更有价值的kmer作为minimizer(when minimizers do not cover all the letters in a string, it is especially important that they cover ‘valuable’ substrings. ) 有价值的(‘valuable’ substring)这里指频率更低,是后续查找和比对更高效的代表序列。
8.Ordering selection?
Object:
1.if a string contains many consecutive zeros (避免全0的kmer被选为minimizer)
2.we want to devise our ordering to increase the chance of rare k-mers being minim- izers, thus increasing the statistical significance of matching minimizers. (尽量让低频率的kmer被选择为minimizer,从而减少之后查找时的比对次数)
mitigate the above effects:
- 选择一个顺序,出现频率最低的字母作为最小(choosing an ordering in which the let- ters that occur least frequently are deemed minimal)
(In DNA sequences, the letters C and G often occur less frequently than A and T. We assign the values 0, 1, 2, 3 to C,A,T,G)
-
?? (changing the ordering from one letter to the next)
-
random order
9.Reverse complement?
Finally, for DNA sequences we are also interested in matches between one string and the reverse complement of another string. Thus in choosing seeds, we identify each k-mer with its reverse complement. Then we choose the minimizer of each window W to be the smaller of the two minimizers from W and its reverse complement.
10.how many memories do all seeds take ?

11.how much space saving?
in general the space savings is about a factor of 2/(w + 1), see section 3
Comments
- sequence comparison, used in many applications.
- overlap determination in genome sequence assembly
- gene finding and comparision
- protein sequence comparision
不同应用场景的本质区别是 pair-wise comparision V.S. multi-pair-wise comparision
2.至少是random order, 评判标准是 density,产生多少个minimizer(与内存和查询速度有关,与匹配准确率有关)
3.minimizer就是MinHash;就是如何sample k-mer的问题, 怎么sample达到内存速度准确的一个平衡
4.minimizer(是按一定的排序规则挑选的)区别于另一种fixed sample的采样方法
5.minimizer自身方法间的区别 就是采样策略的区别, 最常用的是random order.
6.综述目标: 找minimizer不同问题上具体应用的区别